Spatial¶
# !pip install 'lamindb[jupyter,bionty]'
!lamin init --storage ./test-spatial --schema bionty
Show code cell output
→ connected lamindb: testuser1/test-spatial
import lamindb as ln
import bionty as bt
import matplotlib.pyplot as plt
import scanpy as sc
Show code cell output
→ connected lamindb: testuser1/test-spatial
ln.context.uid = "daeFs3PkquDW0000"
ln.context.track()
→ notebook imports: bionty==0.49.0 lamindb==0.76.3 matplotlib==3.9.2 scanpy==1.10.2
→ created Transform('daeFs3PkquDW0000') & created Run('2024-09-02 13:30:14.683157+00:00')
An example spatial dataset¶
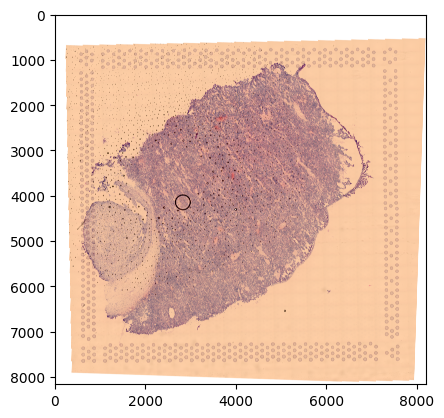
Here, we have a spatial gene expression dataset measured using Visium from Suo22.
This collection contains two parts:
a high-res image of a slice of fetal liver
a single cell expression dataset in .h5ad
img_path = ln.core.datasets.file_tiff_suo22()
img = plt.imread(img_path)
plt.imshow(img)
plt.show()
adata = ln.core.datasets.anndata_suo22_Visium10X()
# subset to the same image
adata = adata[adata.obs["img_id"] == "F121_LP1_4LIV"].copy()
adata
AnnData object with n_obs × n_vars = 3027 × 191
obs: 'in_tissue', 'array_row', 'array_col', 'sample', 'n_genes_by_counts', 'log1p_n_genes_by_counts', 'total_counts', 'log1p_total_counts', 'pct_counts_in_top_50_genes', 'pct_counts_in_top_100_genes', 'pct_counts_in_top_200_genes', 'pct_counts_in_top_500_genes', 'mt_frac', 'img_id', 'EXP_id', 'Organ', 'Fetal_id', 'SN', 'Visium_Area_id', 'Age_PCW', 'Digestion time', 'paths', 'sample_id', '_scvi_batch', '_scvi_labels', '_indices', 'total_cell_abundance'
var: 'feature_types', 'genome', 'SYMBOL', 'mt'
obsm: 'NMF', 'means_cell_abundance_w_sf', 'q05_cell_abundance_w_sf', 'q95_cell_abundance_w_sf', 'spatial', 'stds_cell_abundance_w_sf'
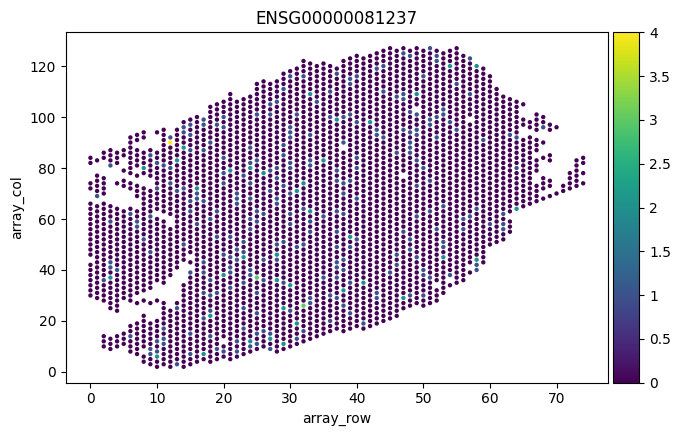
# plot where CD45+ leukocytes are in the slice
sc.pl.scatter(adata, "array_row", "array_col", color="ENSG00000081237")
Validate annotations¶
We’ll register the single-cell data and the image as a Collection.
curate = ln.Curator.from_anndata(adata, var_index=bt.Gene.ensembl_gene_id, categoricals={"sample": ln.ULabel.name}, organism="human")
Show code cell output
✓ added 1 record with Feature.name for columns: 'sample'
• 26 non-validated categories are not saved in Feature.name: ['mt_frac', 'sample_id', 'EXP_id', '_scvi_batch', 'in_tissue', '_scvi_labels', 'Visium_Area_id', 'array_row', 'Organ', 'paths', 'total_cell_abundance', 'pct_counts_in_top_100_genes', 'img_id', 'Fetal_id', 'pct_counts_in_top_50_genes', 'Age_PCW', 'pct_counts_in_top_200_genes', 'array_col', 'log1p_n_genes_by_counts', 'n_genes_by_counts', 'log1p_total_counts', 'pct_counts_in_top_500_genes', 'SN', 'Digestion time', '_indices', 'total_counts']!
→ to lookup categories, use lookup().columns
→ to save, run add_new_from_columns
curate.validate()
✓ created 1 Organism record from Bionty matching name: 'human'
• mapping var_index on Gene.ensembl_gene_id
! found 191 validated terms: ['ENSG00000002586', 'ENSG00000004468', 'ENSG00000004897', 'ENSG00000007312', 'ENSG00000008086', 'ENSG00000008128', 'ENSG00000010278', 'ENSG00000010610', 'ENSG00000012124', 'ENSG00000013725', 'ENSG00000019582', 'ENSG00000026508', 'ENSG00000039068', 'ENSG00000059758', 'ENSG00000062038', 'ENSG00000065883', 'ENSG00000066294', 'ENSG00000070831', 'ENSG00000071991', 'ENSG00000073754', 'ENSG00000074276', 'ENSG00000079112', 'ENSG00000079335', 'ENSG00000081138', 'ENSG00000081237', 'ENSG00000085063', 'ENSG00000085117', 'ENSG00000089486', 'ENSG00000090659', 'ENSG00000091527', 'ENSG00000091972', 'ENSG00000093009', 'ENSG00000094804', 'ENSG00000094880', 'ENSG00000096401', 'ENSG00000097046', 'ENSG00000099804', 'ENSG00000099834', 'ENSG00000100526', 'ENSG00000101017', 'ENSG00000101224', 'ENSG00000101290', 'ENSG00000101391', 'ENSG00000102181', 'ENSG00000102225', 'ENSG00000102245', 'ENSG00000102543', 'ENSG00000103502', 'ENSG00000103855', 'ENSG00000104894', 'ENSG00000105369', 'ENSG00000105383', 'ENSG00000105401', 'ENSG00000105810', 'ENSG00000106993', 'ENSG00000108465', 'ENSG00000110448', 'ENSG00000110651', 'ENSG00000110848', 'ENSG00000111276', 'ENSG00000111328', 'ENSG00000111665', 'ENSG00000112149', 'ENSG00000113100', 'ENSG00000113361', 'ENSG00000114013', 'ENSG00000116031', 'ENSG00000116815', 'ENSG00000116824', 'ENSG00000117091', 'ENSG00000117266', 'ENSG00000117281', 'ENSG00000117335', 'ENSG00000117399', 'ENSG00000117877', 'ENSG00000120217', 'ENSG00000121594', 'ENSG00000122223', 'ENSG00000123080', 'ENSG00000123374', 'ENSG00000124215', 'ENSG00000124762', 'ENSG00000125726', 'ENSG00000125810', 'ENSG00000128283', 'ENSG00000128536', 'ENSG00000129226', 'ENSG00000129355', 'ENSG00000129596', 'ENSG00000129757', 'ENSG00000129910', 'ENSG00000130177', 'ENSG00000132964', 'ENSG00000134058', 'ENSG00000134061', 'ENSG00000134371', 'ENSG00000134690', 'ENSG00000135218', 'ENSG00000135404', 'ENSG00000135446', 'ENSG00000135535', 'ENSG00000136807', 'ENSG00000136861', 'ENSG00000137101', 'ENSG00000138395', 'ENSG00000139193', 'ENSG00000140326', 'ENSG00000140743', 'ENSG00000140937', 'ENSG00000140945', 'ENSG00000143119', 'ENSG00000143776', 'ENSG00000144354', 'ENSG00000145996', 'ENSG00000146670', 'ENSG00000147883', 'ENSG00000147889', 'ENSG00000148600', 'ENSG00000149798', 'ENSG00000150637', 'ENSG00000151465', 'ENSG00000153046', 'ENSG00000153283', 'ENSG00000153563', 'ENSG00000155111', 'ENSG00000156535', 'ENSG00000158402', 'ENSG00000158473', 'ENSG00000158481', 'ENSG00000158485', 'ENSG00000158488', 'ENSG00000158825', 'ENSG00000158985', 'ENSG00000160654', 'ENSG00000163171', 'ENSG00000163606', 'ENSG00000163624', 'ENSG00000163814', 'ENSG00000164045', 'ENSG00000164287', 'ENSG00000164649', 'ENSG00000164885', 'ENSG00000167258', 'ENSG00000167286', 'ENSG00000167513', 'ENSG00000167775', 'ENSG00000167797', 'ENSG00000167850', 'ENSG00000167851', 'ENSG00000168438', 'ENSG00000168564', 'ENSG00000169217', 'ENSG00000169442', 'ENSG00000170312', 'ENSG00000170458', 'ENSG00000170558', 'ENSG00000170779', 'ENSG00000172116', 'ENSG00000173762', 'ENSG00000174059', 'ENSG00000174807', 'ENSG00000176386', 'ENSG00000177455', 'ENSG00000177575', 'ENSG00000177675', 'ENSG00000177697', 'ENSG00000178562', 'ENSG00000178789', 'ENSG00000179604', 'ENSG00000179776', 'ENSG00000184661', 'ENSG00000185324', 'ENSG00000186074', 'ENSG00000186407', 'ENSG00000196352', 'ENSG00000196776', 'ENSG00000197622', 'ENSG00000198087', 'ENSG00000198752', 'ENSG00000198821', 'ENSG00000198851', 'ENSG00000204539', 'ENSG00000204936', 'ENSG00000205643', 'ENSG00000206531', 'ENSG00000213402', 'ENSG00000237190', 'ENSG00000241399', 'ENSG00000248333', 'ENSG00000272398', 'ENSG00000273061']
→ save terms via .add_validated_from_var_index()
! 1 terms is not validated: 'WSSS_F_IMMsp9838712'
→ save terms via .add_new_from('sample')
False
curate.add_validated_from_var_index()
curate.add_new_from('sample')
✓ added 1 record with ULabel.name for sample: 'WSSS_F_IMMsp9838712'
curate.validate()
✓ var_index is validated against Gene.ensembl_gene_id
✓ sample is validated against ULabel.name
True
Register curated artifact¶
artifact_ad = curate.save_artifact(description="Suo22 Visium10X image F121_LP1_4LIV")
Show code cell output
• path content will be copied to default storage upon `save()` with key `None` ('.lamindb/nAGvE7pKiRNgUzdd0000.h5ad')
✓ storing artifact 'nAGvE7pKiRNgUzdd0000' at '/home/runner/work/lamin-usecases/lamin-usecases/docs/test-spatial/.lamindb/nAGvE7pKiRNgUzdd0000.h5ad'
• parsing feature names of X stored in slot 'var'
✓ 191 terms (100.00%) are validated for ensembl_gene_id
✓ linked: FeatureSet(uid='tJ7bTd9WxSPVKSPH5rR8', n=191, dtype='float', registry='bionty.Gene', hash='f29u0HJ47KiqdYQuuhNzeQ', created_by_id=1, run_id=1)
• parsing feature names of slot 'obs'
✓ 1 term (3.70%) is validated for name
! 26 terms (96.30%) are not validated for name: in_tissue, array_row, array_col, n_genes_by_counts, log1p_n_genes_by_counts, total_counts, log1p_total_counts, pct_counts_in_top_50_genes, pct_counts_in_top_100_genes, pct_counts_in_top_200_genes, pct_counts_in_top_500_genes, mt_frac, img_id, EXP_id, Organ, Fetal_id, SN, Visium_Area_id, Age_PCW, Digestion time, ...
✓ linked: FeatureSet(uid='oudlY1aA9YabSBxkUnYf', n=1, registry='Feature', hash='ooDZDOGmzdQLXVBZf6OPhA', created_by_id=1, run_id=1)
✓ saved 2 feature sets for slots: 'var','obs'
artifact_ad.describe()
Artifact(uid='nAGvE7pKiRNgUzdd0000', is_latest=True, description='Suo22 Visium10X image F121_LP1_4LIV', suffix='.h5ad', type='dataset', size=9743793, hash='MRyvckic_gbrV_hHpHOlAQ', n_observations=3027, _hash_type='md5', _accessor='AnnData', visibility=1, _key_is_virtual=True, updated_at='2024-09-02 13:30:29 UTC')
Provenance
.created_by = 'testuser1'
.storage = '/home/runner/work/lamin-usecases/lamin-usecases/docs/test-spatial'
.transform = 'Spatial'
.run = '2024-09-02 13:30:14 UTC'
Labels
.ulabels = 'WSSS_F_IMMsp9838712'
Features
'sample' = 'WSSS_F_IMMsp9838712'
Feature sets
'var' = 'CD99', 'CD38', 'CDC27', 'CD79B', 'CDKL5', 'CDK11A', 'CD9', 'CD4', 'CD22', 'CD6', 'CD74', 'CD44', 'CDH1', 'CDK17', 'CDH3', 'CDK13', 'CD84', 'CDC42', 'CDH19', 'CD5L'
'obs' = 'sample'
Register a collection¶
artifact_img = ln.Artifact(img_path, description="Suo22 image F121_LP1_4LIV")
artifact_img.save()
Show code cell output
• path content will be copied to default storage upon `save()` with key `None` ('.lamindb/zIXAGYoCoEttRSlN0000.tiff')
✓ storing artifact 'zIXAGYoCoEttRSlN0000' at '/home/runner/work/lamin-usecases/lamin-usecases/docs/test-spatial/.lamindb/zIXAGYoCoEttRSlN0000.tiff'
Artifact(uid='zIXAGYoCoEttRSlN0000', is_latest=True, description='Suo22 image F121_LP1_4LIV', suffix='.tiff', size=119764004, hash='ZAnyai4Ys01P2fLR_aDIvq', _hash_type='sha1-fl', visibility=1, _key_is_virtual=True, created_by_id=1, storage_id=1, transform_id=1, run_id=1, updated_at='2024-09-02 13:30:29 UTC')
collection = ln.Collection([artifact_ad, artifact_img], name="Suo22")
collection.save()
Collection(uid='MWvEfPvbxcU6yp1G0000', is_latest=True, name='Suo22', hash='8BBgP1aPBOFv9jCebMhGdw', visibility=1, created_by_id=1, transform_id=1, run_id=1, updated_at='2024-09-02 13:30:30 UTC')
# clean up test instance
!lamin delete --force test-spatial
!rm -r test-flow
Show code cell output
Traceback (most recent call last):
File "/opt/hostedtoolcache/Python/3.10.14/x64/bin/lamin", line 8, in <module>
sys.exit(main())
File "/opt/hostedtoolcache/Python/3.10.14/x64/lib/python3.10/site-packages/rich_click/rich_command.py", line 367, in __call__
return super().__call__(*args, **kwargs)
File "/opt/hostedtoolcache/Python/3.10.14/x64/lib/python3.10/site-packages/click/core.py", line 1157, in __call__
return self.main(*args, **kwargs)
File "/opt/hostedtoolcache/Python/3.10.14/x64/lib/python3.10/site-packages/rich_click/rich_command.py", line 152, in main
rv = self.invoke(ctx)
File "/opt/hostedtoolcache/Python/3.10.14/x64/lib/python3.10/site-packages/click/core.py", line 1688, in invoke
return _process_result(sub_ctx.command.invoke(sub_ctx))
File "/opt/hostedtoolcache/Python/3.10.14/x64/lib/python3.10/site-packages/click/core.py", line 1434, in invoke
return ctx.invoke(self.callback, **ctx.params)
File "/opt/hostedtoolcache/Python/3.10.14/x64/lib/python3.10/site-packages/click/core.py", line 783, in invoke
return __callback(*args, **kwargs)
File "/opt/hostedtoolcache/Python/3.10.14/x64/lib/python3.10/site-packages/lamin_cli/__main__.py", line 179, in delete
return delete(instance, force=force)
File "/opt/hostedtoolcache/Python/3.10.14/x64/lib/python3.10/site-packages/lamindb_setup/_delete.py", line 98, in delete
n_objects = check_storage_is_empty(
File "/opt/hostedtoolcache/Python/3.10.14/x64/lib/python3.10/site-packages/lamindb_setup/core/upath.py", line 776, in check_storage_is_empty
raise InstanceNotEmpty(message)
lamindb_setup.core.upath.InstanceNotEmpty: Storage /home/runner/work/lamin-usecases/lamin-usecases/docs/test-spatial/.lamindb contains 2 objects ('_is_initialized' ignored) - delete them prior to deleting the instance
rm: cannot remove 'test-flow': No such file or directory
